12.5: Try It!
( \newcommand{\kernel}{\mathrm{null}\,}\)
Ginkgo Biloba is recognized as a herbal remedy for memory improvement. To investigate its effectiveness on memory recall, a cross-over study was planned using 3 treatments: one tablet of 120mg Ginkgo Biloba (G), one tablet of 200mg Caffeine pill (C), and sleep for 2 hours before the recall test (S). The assignment order of the 3 treatments to participants was determined by randomly assigning 12 college students to one of 6 possible sequences of the 3 treatments. The student recall capability was assessed based on a Recall score and the 3 treatments were given over 3 consecutive days. On each day, only one treatment was administered before one 1 hour of taking the recall test at 2.00 pm (the higher the recall score the better).
a) Which variable signifies the experimental unit?
- Solution
-
Id
b) What is the washout period?
- Solution
-
One day
c) How many periods are required?
- Solution
-
3
d) How many replicates are there?
- Solution
-
2
e) Perform a statistical analysis to determine if the treatments vary with regard to memory recall. The data can be found in Cross_over_Ex1.txt
- Solution: Using SAS
-
DATA CROSS_OVER; INPUT score Seq $ PER Id TRT $ X1 X2; DATALINES; 74 CGS 1 1 C 0 0 45 CGS 1 2 C 0 0 92 CSG 1 3 C 0 0 94 CSG 1 4 C 0 0 79 GCS 1 5 G 0 0 35 GCS 1 6 G 0 0 31 GSC 1 7 G 0 0 40 GSC 1 8 G 0 0 106 SCG 1 9 S 0 0 60 SCG 1 10 S 0 0 80 SGC 1 11 S 0 0 110 SGC 1 12 S 0 0 41 CGS 2 1 G 1 0 20 CGS 2 2 G 1 0 50 CSG 2 3 S 1 0 88 CSG 2 4 S 1 0 92 GCS 2 5 C 0 1 50 GCS 2 6 C 0 1 32 GSC 2 7 S 0 1 54 GSC 2 8 S 0 1 120 SCG 2 9 C -1 -1 80 SCG 2 10 C -1 -1 75 SGC 2 11 G -1 -1 55 SGC 2 12 G -1 -1 64 CGS 3 1 S 0 1 30 CGS 3 2 S 0 1 55 CSG 3 3 G -1 -1 55 CSG 3 4 G -1 -1 76 GCS 3 5 S 1 0 50 GCS 3 6 S 1 0 38 GSC 3 7 C -1 -1 66 GSC 3 8 C -1 -1 85 SCG 3 9 G 1 0 40 SCG 3 10 G 1 0 88 SGC 3 11 C 0 1 86 SGC 3 12 C 0 1 ; RUN; proc mixed data=CROSS_OVER; class PER TRT SEQ ID; model SCORE=PER TRT SEQ X1 X2 / ddfm=kr; repeated PER /subject=ID(SEQ) type=cs rcorr; ods output FitStatistics=FitCS (rename=(value=CS)) FitStatistics=FitCSp; title 'Compound Symmetry'; run; title ' '; run; proc mixed data=CROSS_OVER; class PER TRT SEQ ID; model SCORE=PER TRT SEQ X1 X2 / ddfm=kr; repeated PER /subject=ID(SEQ) type=AR(1) rcorr; ods output FitStatistics=FitAR1 (rename=(value=AR1)) FitStatistics=FitAR1p; title 'Autoregressive Lag 1'; run; title ' '; run; proc mixed data=CROSS_OVER; class PER TRT SEQ ID; model SCORE=PER TRT SEQ X1 X2 / ddfm=kr; repeated PER /subject=ID(SEQ) type=UN rcorr; ods output FitStatistics=FitUN (rename=(value=UN)) FitStatistics=FitUNp; title 'Unstructured'; run; title ' '; run; proc mixed data=CROSS_OVER; class PER TRT SEQ ID; model SCORE=PER TRT SEQ X1 X2 / ddfm=kr; repeated PER /subject=ID(SEQ) type=CSH rcorr; ods output FitStatistics=FitCSH (rename=(value=CSH)) FitStatistics=FitCSHp; title 'HETEROGENOUS COMPOUND SYMMETRY'; run; title ' '; run; data fits; merge FitCS FitAR1 FitUN FITCSH; by descr; run; ods listing; proc print data=fits; run;
The above code was used to obtain the fit statistics for different covariance structures and the AICC (AIC and BIC) values indicate that CS is the best covariance structure. Hence, the remaining analysis was done using CS.
Obs Descr CS AR1 UN CSH 1 -2 Res Log Likelihood 215.3 219.1 212.7 214.7 2 AIC (Smaller is Better) 219.3 223.1 224.7 222.7 3 AICC (Smaller is Better) 219.9 223.7 229.7 224.8 4 BIC (Smaller is Better) 220.3 224.1 227.6 224.6 /* Model Adjusting for carryover effects */ proc mixed data= CROSS_OVER; class per TRT SEQ ID; model SCORE=PER TRT SEQ X1 X2 / ddfm=kr; repeated PER /subject=ID(SEQ) type=cs rcorr; store out_CROSS_OVER; run; proc plm restore=out_CROSS_OVER; lsmeans TRT / adjust=tukey plot=meanplot cl lines; ods exclude diffs diffplot; run; /* Reduced Model, Ignoring carryover effects */ proc mixed data= CROSS_OVER; class per TRT seq ID; model SCORE=PER TRT SEQ / ddfm=kr; repeated PER /subject=ID(SEQ) type=cs rcorr; lsmeans TRT / pdiff adjust=tukey; run;
Full Model: with carryover effect
Fit Statistics -2 Res Log Likelihood 215.3 AIC (Smaller is Better) 219.3 AICC (Smaller is Better) 219.9 BIC (Smaller is Better) 220.3 Type 3 Tests of Fixed Effects Effect Num DF Den DF F Value Pr > F PER 2 18 3.12 0.0688 TRT 2 18 18.03 <.0001 Seq 5 6.1 1.46 0.3259 X1 1 18 0.10 0.7565 X2 1 18 0.18 0.6768 Reduced Model: without carryover effect
Fit Statistics -2 Res Log Likelihood 224.2 AIC (Smaller is Better) 228.2 AICC (Smaller is Better) 228.7 BIC (Smaller is Better) 229.1 Type 3 Tests of Fixed Effects Effect Num DF Den DF F Value Pr > F PER 2 20 3.36 0.0552 TRT 2 20 23.70 <.0001 Seq 5 6 1.52 0.3101 The test statistic below tests for the significance of the carry over effect. Δ2=(−2logLReduced)−(−2logLFull)with\(dfFull−dfReduced degrees of freedom.
Δ2=(224.2−215.3)=8.9. This exceeds the critical Chi-Square of 5.991 (χ2.05,2) indicating that model with carryover effect is more appropriate and will be used to base the final conclusions.
In the full model output, the Treatment is the only significant factor, so LSmeans and comparisons are generated only for the treatment effect. The results of the Tukey comparison procedure indicate that treatments C and S are not significantly different, but G is significantly lower, indicating that both sleep for 2 hours and caffeine are similarly effective in improving recall capability and are superior to Ginkgo biloba.
TRT Least Squares Means TRT Estimate Standard Error DF t Value Pr > |t| Alpha Lower Upper C 76.7222 6.2382 8.572 12.30 <.0001 0.05 62.5024 90.9421 G 50.4306 6.2382 8.572 8.08 <.0001 0.05 36.2107 64.6504 S 67.5139 6.2382 8.572 10.82 <.0001 0.05 53.2940 81.7337 -
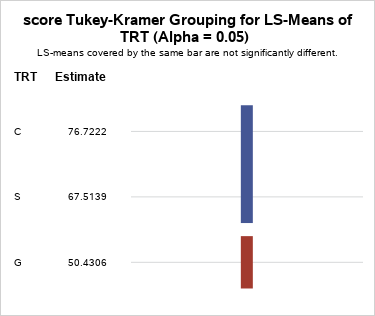
Figure 12.5.1: Scores for Tukey-Kramer grouping


