3.7: One-Way ANOVA Greenhouse Example in R
- Page ID
- 33440
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)R Instructions: Code for the Greenhouse Data
- Load the greenhouse data.
- Calculate the overall mean, standard deviation, and standard error.
- Calculate the mean, standard deviation, and standard error for each group.
- Produce a boxplot to plot the differences in heights for each fertilizer.
- Produce a "means plot" (interval plot) to view the differences in heights for each fertilizer.
- Obtain the ANOVA table.
- Obtain Tukey’s multiple comparisons CIs and difference in means plot.
- Produce diagnostic (residuals) plots.
- Power analysis.
setwd("~/path-to-folder/")
greenhouse_data<-read.table("greenhouse_data.txt",header=T)
Note that greenhouse data are in separate columns.
attach(greenhouse_data) my_data<-stack(greenhouse_data)
With this command, we put our data in a stacked format (the first column has the response variable (values) and the second column has the treatment levels (ind).
To calculate the overall mean, standard deviation, and standard error we can use the following commands:
overall_mean<-mean(my_data$values) #26.16667 overall_sd<-sd(my_data$values) #3.685892 overall_standard_error<-overall_sd/sqrt(length(my_data$values)) #0.7523795
To calculate the group means we can use the following command:
group_means<-aggregate(my_data[, 1],list(my_data$ind),mean)
# group_means
# Group.1 x
# 1 Control 21.00000
# 2 F1 28.60000
# 3 F2 25.86667
# 4 F3 29.20000
To calculate the group standard deviations and standard errors we can use the following commands:
group_sd<-aggregate(my_data[, 1],list(my_data$ind),sd)
# group_sd Group.1 x
# 1 Control 1.000000
# 2 F1 2.437212
# 3 F2 1.899123
# 4 F3 1.288410
group_standard_error<-group_sd$x/sqrt(length(my_data$ind)/4)
# group_standard_error
# 0.4082483 0.9949874 0.7753136 0.5259911
To produce the Boxplot we can use the following commands:
library("ggpubr")
boxplot(values~ind,data=my_data,
xlab="Fertilizer",ylab="Plant Height",
main="Distribution of Plant Heights by Fertilizer",
frame=TRUE)
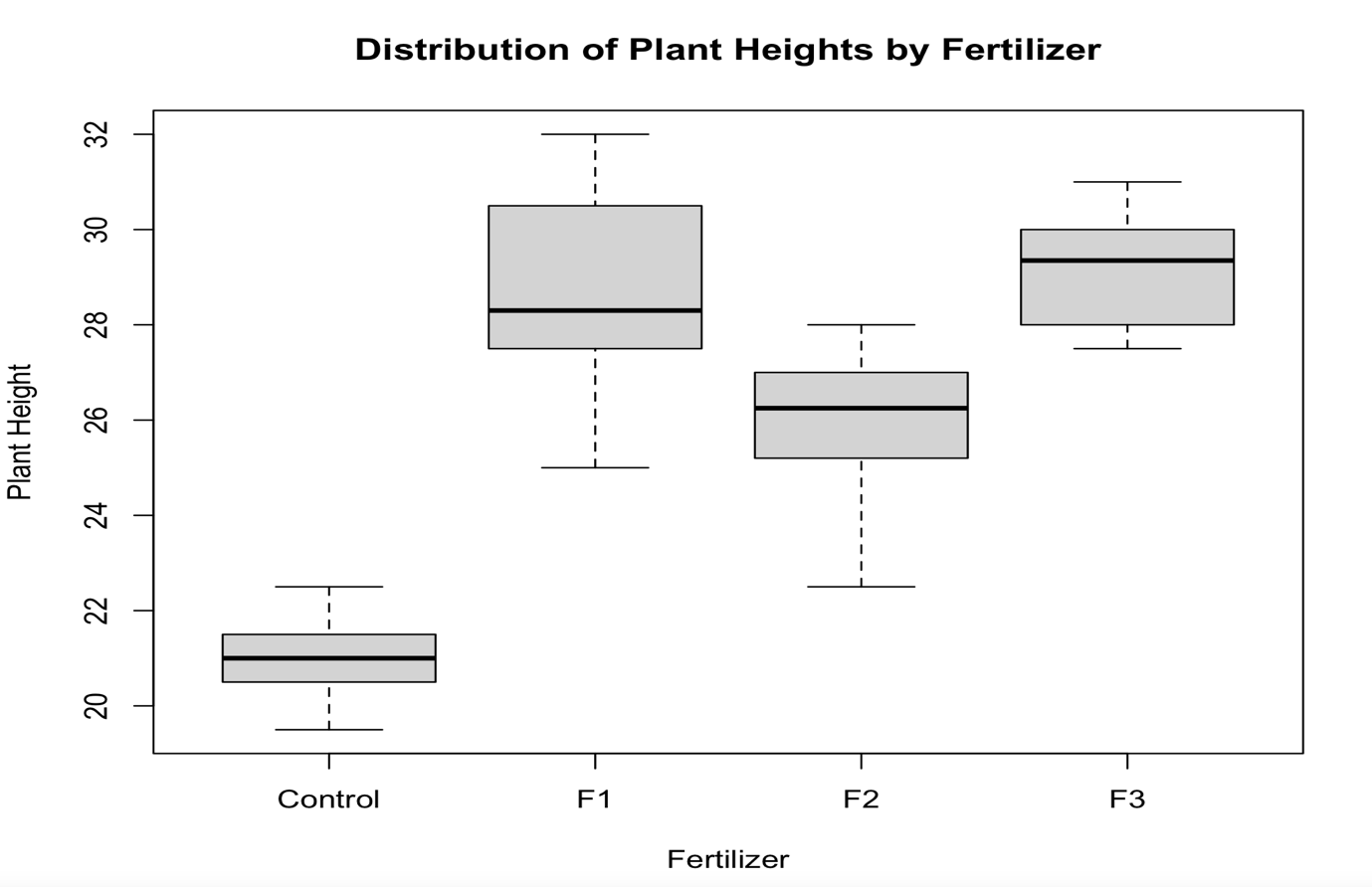
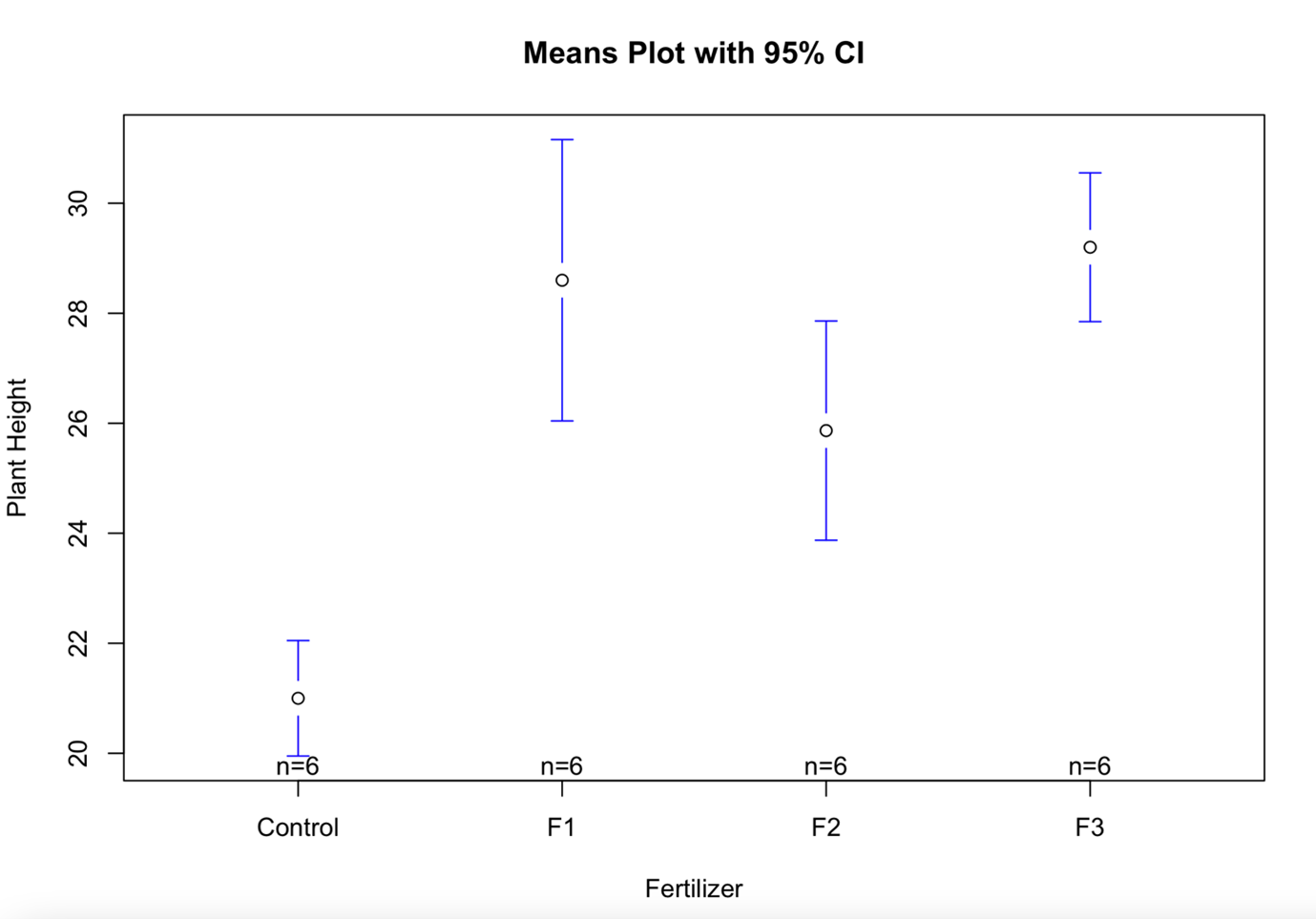
To produce the means plot (interval plot) we can use the following commands:
library("gplots")
plotmeans(values~ind,data=my_data,connect=FALSE,
xlab="Fertilizer",ylab="Plant Height",
main="Means Plot with 95% CI")
To obtain the ANOVA table we can use the following commands:
anova<-aov(values~ind,my_data) summary(anova)
The command summary (anova) will give you the following output:
summary(anova) Df Sum Sq Mean Sq F value Pr(>F) ind 3 251.44 83.81 27.46 2.71e-07 *** Residuals 20 61.03 3.05 --- Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
We can see the degrees of freedom in the first column, the sum of squares in the second column, the mean sum of squares in the third column, the \(F\)-test statistic in the fourth column, and finally, we can see the \(p\)-value.
Note that the output doesn't give the \(SSTO\). To find it, use the identity \(SSTO=SSR+SSE\). Similarly, for the \(df\) associated with \(SSTO\), add the \(df\) of \(SSR\) and \(SSE\).
For our example, \(SSTO=251.44+61.03=312.47\)
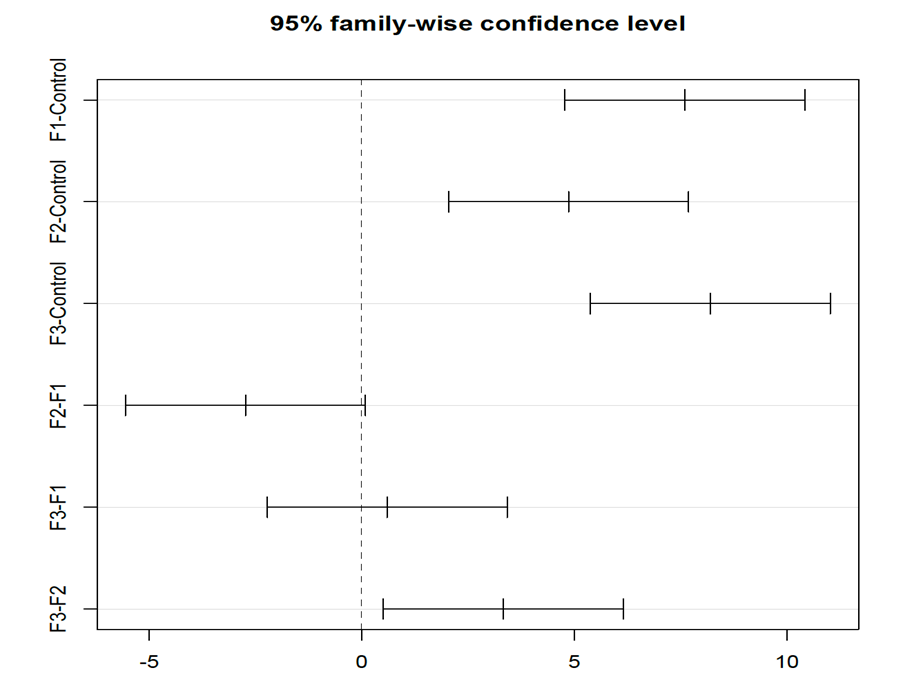
To obtain Tukey multiple comparisons of means with a 95% family-wise confidence level we use the following command:
library(multcomp) library(multcompView) tukey_multiple_comparisons<-TukeyHSD(anova,conf.level=0.95) plot(tukey_multiple_comparisons) tukey_multiple_comparisons Tukey multiple comparisons of means 95% family-wise confidence level Fit: aov(formula = values ~ ind, data = my_data) $ind diff lwr upr p adj F1-Control 7.600000 4.7770648 10.42293521 0.0000016 F2-Control 4.866667 2.0437315 7.68960188 0.0005509 F3-Control 8.200000 5.3770648 11.02293521 0.0000005 F2-F1 -2.733333 -5.5562685 0.08960188 0.0598655 F3-F1 0.600000 -2.2229352 3.42293521 0.9324380 F3-F2 3.333333 0.5103981 6.15626854 0.0171033
Based on this output, the Control group is significantly different from the 3 treatment groups and F3 is significantly different from F2.
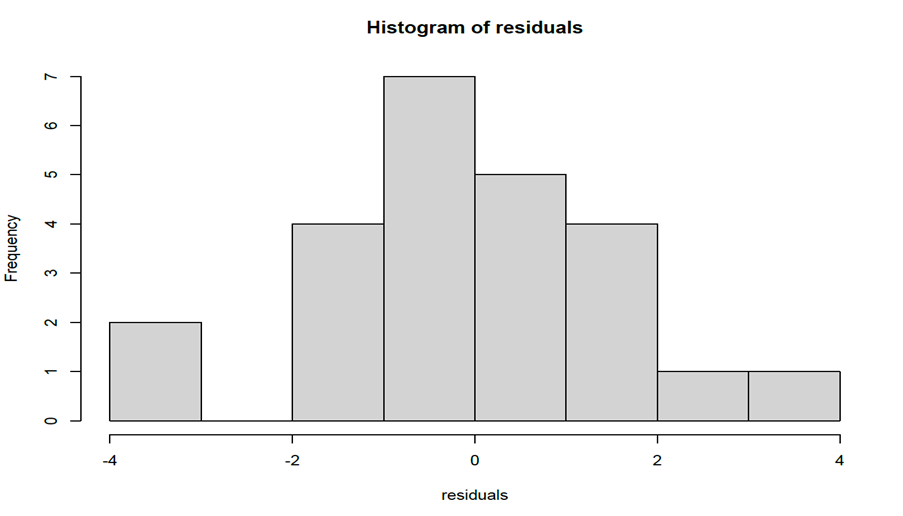
To produce diagnostic (residuals) plots we use the following commands:
#Residuals vs Fits plot
plot(anova,1)
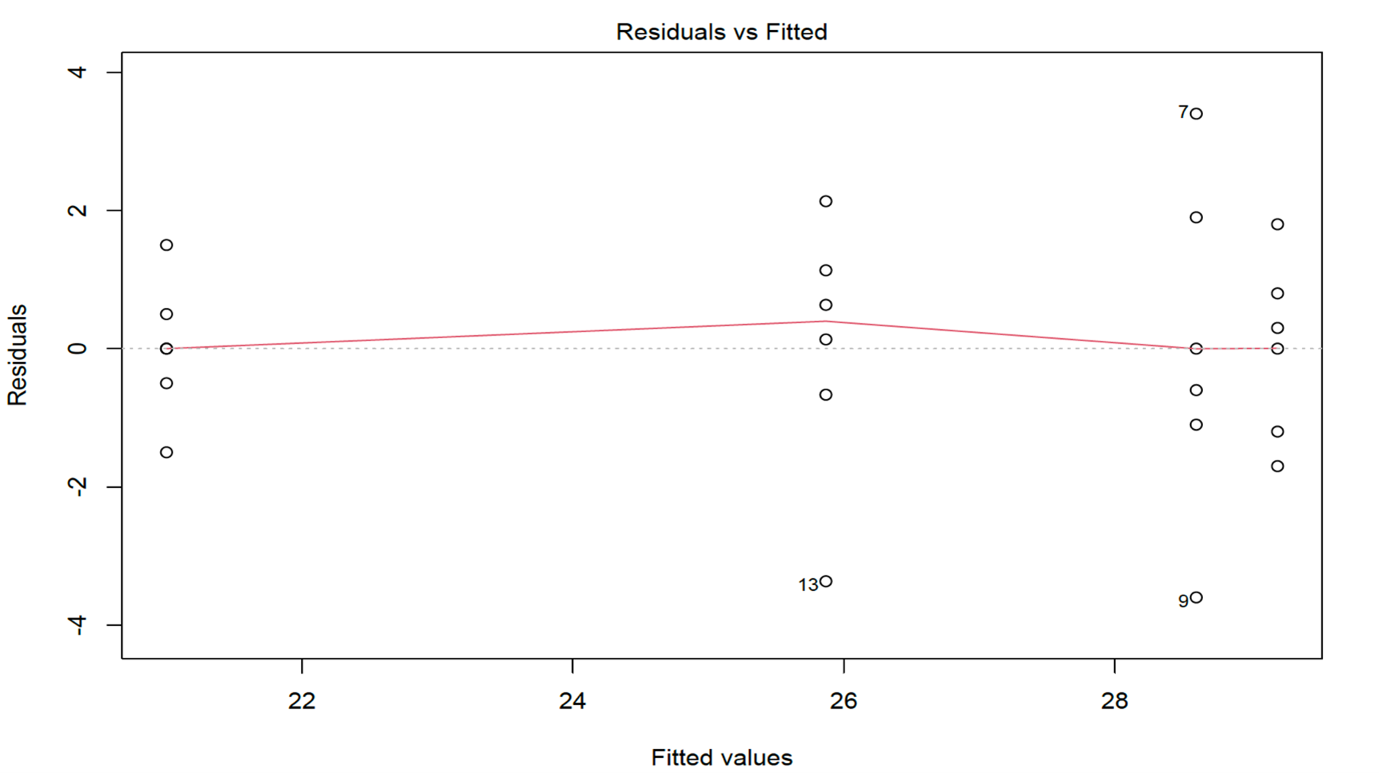
#QQ plot
plot(anova,2)
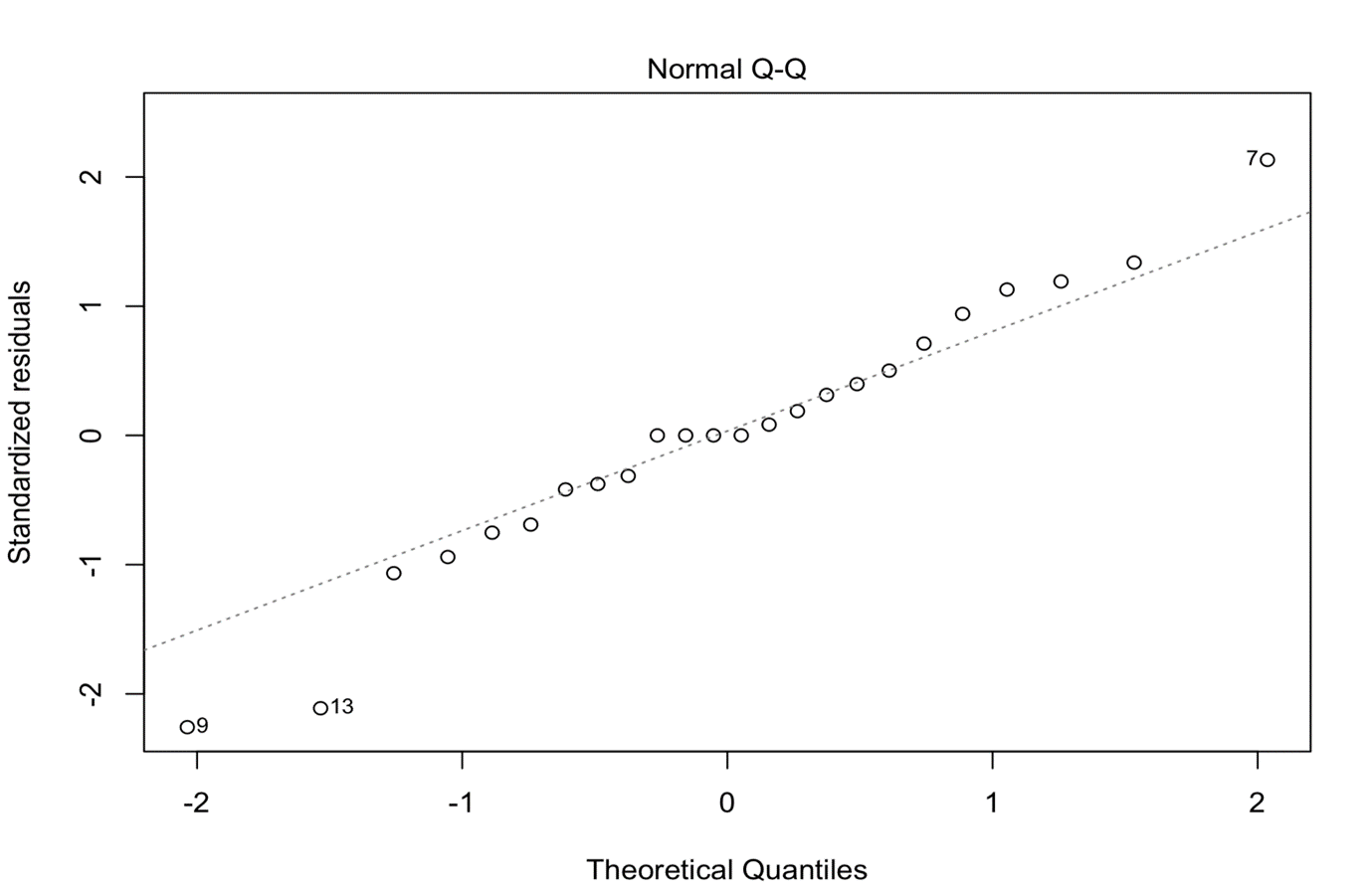
#Histogram of residuals residuals<-anova$res #with this command we get the residuals from ANOVA hist(residuals)