8.1: Split-Plot Design in RCBD
- Page ID
- 33901
Recall the Randomized Complete Block Design (RCBD) we discussed in Chapter 7. In RCBD, general blocks are formed such that the experimental units are expected to be homogenous within a block and heterogeneous between blocks.
For example. suppose we are studying the effect of irrigation amount (\(I_{1}\) and \(I_{2}\)) and fertilizer type (\(A\) and \(B\)) on crop yield. We have 4 treatments in this experiment. Suppose we want to have at least 2 replicates and have two large lands that can be used for the experiment. In RCBD, we can split each land into 4 fields and can apply the 4 treatments randomly to each field. Here lands are blocks and fields are the experimental units.

In this example, we have assumed that managing levels of irrigation and fertilizer require the same effort. Now suppose varying the level of irrigation is difficult on a small scale and it makes more sense to apply irrigation levels to larger areas of land.
In such situations, we can divide each land into two large fields (whole plots) and apply irrigation amounts to each field randomly. And then divide each of these large fields into smaller fields (subplots) and apply fertilizer randomly within the whole plots.
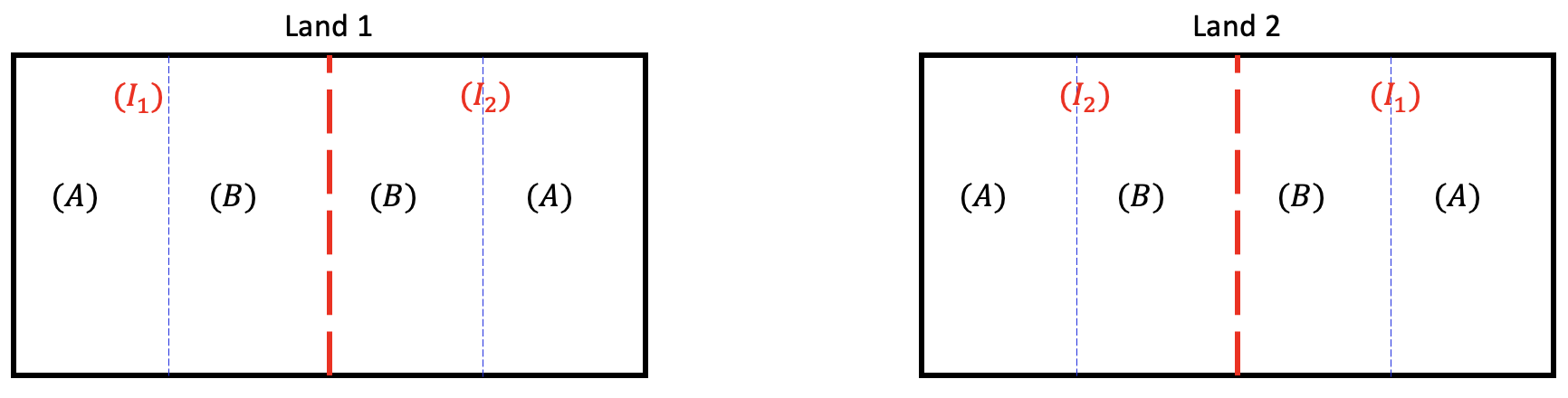
In this strategy, each land contains two whole plots and irrigation amount is assigned to each whole plot randomly using RCBD (i.e. lands are treated as blocks and irrigation amount is assigned randomly within each block to the whole plots). Each whole plot contains two subplots and fertilizer type is assigned to each subplot using RCBD (i.e. whole plots are treated as blocks and fertilizer type is assigned randomly within each whole plot to the subplots).
When some factors are more difficult to vary than others at the levels of experimental units, it is more efficient to assign more difficult-to-change factors to larger units (whole plots) and then apply the easier-to-change factor to smaller units (subplots). This is known as the split-plot design.
As an example (adapted from Hicks, 1964), consider an experiment where an electrical component is subjected to 4 different temperatures for 3 different amounts of time. If the investigators desire 3 replications for each of the 12 temperature and time combinations (i.e. 12 treatments), a basic CRD or an RCBD (with a suitable blocking factor that would generate the replicates) will require as many as 36 attempts of testing.
Instead, the experimentation can be modified as follows to reduce effort and time. Regarding ovens as blocks, 3 ovens can be set to each of the 4 different temperature settings and then investigators can take out randomly selected components at the 3 different times of interest.
In this setting, temperatures are assigned randomly within each oven (i.e. an oven is treated as a block) and within each temperature, the baking times are assigned randomly to components. We have two RCBD sub-experiments: whole plot levels (temperatures) are assigned as RCBD within the oven and subplots levels (baking time) are assigned as RCBD within whole plot levels.
The data (Bake Time Data) were:
| Oven Temperature \((^{\circ} \mathrm{F})\) | |||||
|---|---|---|---|---|---|
| Rep | Baking Time (min) | 580 | 600 | 620 | 640 |
| I | 5 | 217 | 158 | 229 | 223 |
| 10 | 233 | 138 | 186 | 227 | |
| 15 | 175 | 152 | 155 | 156 | |
| II | 5 | 188 | 126 | 160 | 201 |
| 10 | 201 | 130 | 170 | 181 | |
| 15 | 195 | 147 | 161 | 172 | |
| III | 5 | 162 | 122 | 167 | 182 |
| 10 | 170 | 185 | 181 | 201 | |
| 15 | 213 | 180 | 182 | 199 | |
It is important to notice that in a split-plot design, randomization is a two-stage process. Levels of one factor (say, factor A) are randomized over the whole plots within each block, and the levels of the other factor (say, factor B) are randomized over the subplots within each whole plot. This restriction in randomization results in two different error terms: one appropriate for comparisons at the whole plot level and one appropriate for comparisons at the subplot level.
The appropriate error for whole plot level in split-plot RCBD is \(\text{whole plot factor} \times \text{block interaction}\). In other words, the analysis at the whole plot level is essentially of a one-way ANOVA with blocking (i.e. one observation per block-treatment combination). From the perspective of the whole plot, the subplots are simply subsamples and it is reasonable to average them when testing the whole plot effects (i.e. factor A effects).
The subplot factor (i.e. factor B) is always compared within the whole plot factor.
| Source | DF |
|---|---|
| Blocks | \(r-1\) |
| Factor \(A\) | \(a-1\) |
| Whole plot Error | \((r-1)(a-1)\) |
| Factor B | \(b-1\) |
| \(A \times B\) | \((a-1)(b-1)\) |
| Subplot Error | \(a(r-1)(b-1)\) |
| Total | \(rab-1\) |
The statistical model associated with the split-plot design with whole plots arranged as RCBD is \[Y_{ijk} = \mu + \alpha_{i} + \gamma_{k} + (\alpha \gamma)_{ik} + \beta_{j} + (\alpha \beta)_{ij} + \epsilon_{ijk}\] where \(\gamma_{k}\) for \(k=1,\ldots,r\) are block effects, \(\alpha_{i}\) for \(i=1,\ldots,a\) are factor A effects, and \(\beta_{j}\) for \(j=1,\ldots,b\) are factor B effects.
Using Technology
- Steps in SAS
-
In SAS, we could specify the model with the following statements:
proc mixed data=BakeTimeData method=type3; class oven temp time; model resp=temp time temp*time; random oven oven*temp; run;
This will generate the ANOVA table as shown below.
Type 3 Analysis of Variance Source DF Sum of Squares Mean Square Expected Mean Square Error Term Error DF F Value Pr > F temp 3 12494 4164.768519 Var(Residual) + 3 Var(oven*temp) + Q(temp,temp*time) MS(oven*temp) 6 14.09 0.0040 time 2 566.222222 283.111111 Var(Residual) + Q(time,temp*time) MS(Residual) 16 0.46 0.6418 temp*time 6 2600.444444 433.407407 Var(Residual) + Q(temp*time) MS(Residual) 16 0.70 0.6551 oven 2 1962.722222 981.361111 Var(Residual) + 3 Var(oven*temp) + 12 Var(oven) MS(oven*temp) 6 3.32 0.1070 oven*temp 6 1773.944444 295.657407 Var(Residual) + 3 Var(oven*temp) MS(Residual) 16 0.48 0.8162 Residual 16 9933.333333 620.833333 Var(Residual) . . . . The ANOVA table can be rearranged to the following to make it easier to understand the whole plot and subplot analyses.
Source DF Expected Mean Square (Whole Plots) oven 2 Var(Residual) + 3 Var(block*temp) + 12 Var(oven) temp 3 Var(Residual) + 3 Var(oven*temp) + Q(temp, temp*time) oven*temp 6 Var(Residual) + 3 Var(oven*temp) (Subplots) time 2 Var(Residual) + Q(time, temp*time) temp*time 6 Var(Residual) + Q(temp*time) Residual 16 Var(Residual) Notice that the correct error term for the \(F\)-test of the treatment applied to whole plots is the \(\text{block} \times \text{whole plot factor}\) (assuming blocks are a random effect).
One might wonder about the terms \(\text{block} \times \text{subplot factor}\) and \(\text{block} \times \text{whole plot factor} \times \text{subplot factor}\). With these terms in the model, we will not be able to retrieve the residual (the error DF will be zero). If repeat observations are made within the split-plots, then a separate error term can be estimated. However, it is important to keep in mind that tests of replication effects are not of interest, but are being isolated in the ANOVA to reduce the error variance. As a result, the model that is usually run in this design drops out the \(\text{block} \times \text{subplot factor}\) and \(\text{block} \times \text{whole plot factor} \times \text{subplot factor}\) terms, and combine these interactions with the true error variance to obtain a working error term.
- Steps in R
-
Load the bake time data and obtain the ANOVA table by using the following commands:
setwd("~/path-to-folder/") baketime_data <- read.table("baketime_data.txt",header=T) attach(baketime_data) baketime_anova<-aov(resp ~ factor(temp) + factor(time) + factor(temp):factor(time) + Error(factor(oven)+factor(oven):factor(temp)),baketime_data) summary(baketime_anova) #Error: factor(oven) # Df Sum Sq Mean Sq F value Pr(>F) #Residuals 2 1963 981.4 #Error: factor(oven):factor(temp) # Df Sum Sq Mean Sq F value Pr(>F) #factor(temp) 3 12494 4165 14.09 0.004 ** #Residuals 6 1774 296 #--- #Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 #Error: Within # #Df Sum Sq Mean Sq F value Pr(>F) #factor(time) 2 566 283.1 0.456 0.642 #factor(temp):factor(time) 6 2600 433.4 0.698 0.655 #Residuals 16 9933 620.8 detach(baketime_data)


