5.7: Data Cleaning and Tidying with R
- Page ID
- 8732
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Now that you know a bit about the tidyverse, let’s look at the various tools that it provides for working with data. We will use as an example an analysis of whether attitudes about statistics are different between the different student year groups in the class.
5.7.1 Statistics attitude data from course survey
These data were collected using the Attitudes Towards Statistics (ATS) scale (from https://www.stat.auckland.ac.nz/~iase/cblumberg/wise2.pdf).
The 29-item ATS has two subscales. The Attitudes Toward Field subscale consists of the following 20 items, with reverse-keyed items indicated by an “(R)”: 1, 3, 5, 6(R), 9, 10(R), 11, 13, 14(R), 16(R), 17, 19, 20(R), 21, 22, 23, 24, 26, 28(R), 29
The Attitudes Toward Course subscale consists of the following 9 items: 2(R), 4(R), 7(R), 8, 12(R), 15(R), 18(R), 25(R), 27(R)
For our purposes, we will just combine all 29 items together, rather than separating them into these subscales.
Note: I have removed the data from the graduate students and 5+ year students, since those would be too easily identifiable given how few there are.
Let’s first save the file path to the data.
attitudeData_file <- 'data/statsAttitude.txt'Next, let’s load the data from the file using the tidyverse function read_tsv(). There are several functions available for reading in different file formats as part of the the readr tidyverse package.
attitudeData <- read_tsv(attitudeData_file)
glimpse(attitudeData)## Observations: 148
## Variables: 31
## $ `What year are you at Stanford?` <dbl> …
## $ `Have you ever taken a statistics course before?` <chr> …
## $ `I feel that statistics will be useful to me in my profession.` <dbl> …
## $ `The thought of being enrolled in a statistics course makes me nervous.` <dbl> …
## $ `A good researcher must have training in statistics.` <dbl> …
## $ `Statistics seems very mysterious to me.` <dbl> …
## $ `Most people would benefit from taking a statistics course.` <dbl> …
## $ `I have difficulty seeing how statistics relates to my field of study.` <dbl> …
## $ `I see being enrolled in a statistics course as a very unpleasant experience.` <dbl> …
## $ `I would like to continue my statistical training in an advanced course.` <dbl> …
## $ `Statistics will be useful to me in comparing the relative merits of different objects, methods, programs, etc.` <dbl> …
## $ `Statistics is not really very useful because it tells us what we already know anyway.` <dbl> …
## $ `Statistical training is relevant to my performance in my field of study.` <dbl> …
## $ `I wish that I could have avoided taking my statistics course.` <dbl> …
## $ `Statistics is a worthwhile part of my professional training.` <dbl> …
## $ `Statistics is too math oriented to be of much use to me in the future.` <dbl> …
## $ `I get upset at the thought of enrolling in another statistics course.` <dbl> …
## $ `Statistical analysis is best left to the "experts" and should not be part of a lay professional's job.` <dbl> …
## $ `Statistics is an inseparable aspect of scientific research.` <dbl> …
## $ `I feel intimidated when I have to deal with mathematical formulas.` <dbl> …
## $ `I am excited at the prospect of actually using statistics in my job.` <dbl> …
## $ `Studying statistics is a waste of time.` <dbl> …
## $ `My statistical training will help me better understand the research being done in my field of study.` <dbl> …
## $ `One becomes a more effective "consumer" of research findings if one has some training in statistics.` <dbl> …
## $ `Training in statistics makes for a more well-rounded professional experience.` <dbl> …
## $ `Statistical thinking can play a useful role in everyday life.` <dbl> …
## $ `Dealing with numbers makes me uneasy.` <dbl> …
## $ `I feel that statistics should be required early in one's professional training.` <dbl> …
## $ `Statistics is too complicated for me to use effectively.` <dbl> …
## $ `Statistical training is not really useful for most professionals.` <dbl> …
## $ `Statistical thinking will one day be as necessary for efficient citizenship as the ability to read and write.` <dbl> …Right now the variable names are unwieldy, since they include the entire name of the item; this is how Google Forms stores the data. Let’s change the variable names to be somewhat more readable. We will change the names to “atsrename() and paste0() functions. rename() is pretty self-explanatory: a new name is assigned to an old name or a column position. The paste0() function takes a string along with a set of numbers, and creates a vector that combines the string with the number.
nQuestions <- 29 # other than the first two columns, the rest of the columns are for the 29 questions in the statistics attitude survey; we'll use this below to rename these columns based on their question number
# use rename to change the first two column names
# rename can refer to columns either by their number or their name
attitudeData <-
attitudeData %>%
rename( # rename using columm numbers
# The first column is the year
Year = 1,
# The second column indicates whether the person took stats before
StatsBefore = 2
) %>%
rename_at(
# rename all the columns except Year and StatsBefore
vars(-Year, -StatsBefore),
#rename by pasting the word "stat" and the number
funs(paste0('ats', 1:nQuestions))
)
# print out the column names
names(attitudeData)## [1] "Year" "StatsBefore" "ats1" "ats2" "ats3"
## [6] "ats4" "ats5" "ats6" "ats7" "ats8"
## [11] "ats9" "ats10" "ats11" "ats12" "ats13"
## [16] "ats14" "ats15" "ats16" "ats17" "ats18"
## [21] "ats19" "ats20" "ats21" "ats22" "ats23"
## [26] "ats24" "ats25" "ats26" "ats27" "ats28"
## [31] "ats29"#check out the data again
glimpse(attitudeData)## Observations: 148
## Variables: 31
## $ Year <dbl> 3, 4, 2, 1, 2, 3, 4, 2, 2, 2, 4, 2, 3…
## $ StatsBefore <chr> "Yes", "No", "No", "Yes", "No", "No",…
## $ ats1 <dbl> 6, 4, 6, 3, 7, 4, 6, 5, 7, 5, 5, 4, 2…
## $ ats2 <dbl> 1, 5, 5, 2, 7, 5, 5, 4, 2, 2, 3, 3, 7…
## $ ats3 <dbl> 7, 6, 5, 7, 2, 4, 7, 7, 7, 5, 6, 5, 7…
## $ ats4 <dbl> 2, 5, 5, 2, 7, 3, 3, 4, 5, 3, 3, 2, 3…
## $ ats5 <dbl> 7, 5, 6, 7, 5, 4, 6, 6, 7, 5, 3, 5, 4…
## $ ats6 <dbl> 1, 4, 5, 2, 2, 4, 2, 3, 1, 2, 2, 3, 1…
## $ ats7 <dbl> 1, 4, 3, 2, 4, 4, 2, 2, 3, 2, 4, 2, 4…
## $ ats8 <dbl> 2, 1, 4, 3, 1, 4, 4, 4, 7, 3, 2, 4, 1…
## $ ats9 <dbl> 5, 4, 5, 5, 7, 4, 5, 5, 7, 6, 3, 5, 5…
## $ ats10 <dbl> 2, 3, 2, 2, 1, 4, 2, 2, 1, 3, 3, 1, 1…
## $ ats11 <dbl> 6, 4, 6, 2, 7, 4, 6, 5, 7, 3, 3, 4, 2…
## $ ats12 <dbl> 2, 4, 1, 2, 5, 7, 2, 1, 2, 4, 4, 2, 4…
## $ ats13 <dbl> 6, 4, 5, 5, 7, 3, 6, 6, 7, 5, 2, 5, 1…
## $ ats14 <dbl> 2, 4, 3, 3, 3, 4, 2, 1, 1, 3, 3, 2, 1…
## $ ats15 <dbl> 2, 4, 3, 3, 5, 6, 3, 4, 2, 3, 2, 4, 3…
## $ ats16 <dbl> 1, 3, 2, 5, 1, 5, 2, 1, 2, 3, 2, 2, 1…
## $ ats17 <dbl> 7, 7, 5, 7, 7, 4, 6, 6, 7, 6, 6, 7, 4…
## $ ats18 <dbl> 2, 5, 4, 5, 7, 4, 2, 4, 2, 5, 2, 4, 6…
## $ ats19 <dbl> 3, 3, 4, 3, 2, 3, 6, 5, 7, 3, 3, 5, 2…
## $ ats20 <dbl> 1, 4, 1, 2, 1, 4, 2, 2, 1, 2, 3, 2, 3…
## $ ats21 <dbl> 6, 3, 5, 5, 7, 5, 6, 5, 7, 3, 4, 6, 6…
## $ ats22 <dbl> 7, 4, 5, 6, 7, 5, 6, 5, 7, 5, 5, 5, 5…
## $ ats23 <dbl> 6, 4, 6, 6, 7, 5, 6, 7, 7, 5, 3, 5, 3…
## $ ats24 <dbl> 7, 4, 4, 6, 7, 5, 6, 5, 7, 5, 5, 5, 3…
## $ ats25 <dbl> 3, 5, 3, 3, 5, 4, 3, 4, 2, 3, 3, 2, 5…
## $ ats26 <dbl> 7, 4, 5, 6, 2, 4, 6, 5, 7, 3, 4, 4, 2…
## $ ats27 <dbl> 2, 4, 2, 2, 4, 4, 2, 1, 2, 3, 3, 2, 1…
## $ ats28 <dbl> 2, 4, 3, 5, 2, 3, 3, 1, 1, 4, 3, 2, 2…
## $ ats29 <dbl> 4, 4, 3, 6, 2, 1, 5, 3, 3, 3, 2, 3, 2…The next thing we need to do is to create an ID for each individual. To do this, we will use the rownames_to_column() function from the tidyverse. This creates a new variable (which we name “ID”) that contains the row names from the data frame; thsee are simply the numbers 1 to N.
# let's add a participant ID so that we will be able to identify them later
attitudeData <-
attitudeData %>%
rownames_to_column(var = 'ID')
head(attitudeData)## # A tibble: 6 x 32
## ID Year StatsBefore ats1 ats2 ats3 ats4 ats5
## <chr> <dbl> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 1 3 Yes 6 1 7 2 7
## 2 2 4 No 4 5 6 5 5
## 3 3 2 No 6 5 5 5 6
## 4 4 1 Yes 3 2 7 2 7
## 5 5 2 No 7 7 2 7 5
## 6 6 3 No 4 5 4 3 4
## # … with 24 more variables: ats6 <dbl>, ats7 <dbl>,
## # ats8 <dbl>, ats9 <dbl>, ats10 <dbl>, ats11 <dbl>,
## # ats12 <dbl>, ats13 <dbl>, ats14 <dbl>, ats15 <dbl>,
## # ats16 <dbl>, ats17 <dbl>, ats18 <dbl>, ats19 <dbl>,
## # ats20 <dbl>, ats21 <dbl>, ats22 <dbl>, ats23 <dbl>,
## # ats24 <dbl>, ats25 <dbl>, ats26 <dbl>, ats27 <dbl>,
## # ats28 <dbl>, ats29 <dbl>If you look closely at the data, you can see that some of the participants have some missing responses. We can count them up for each individual and create a new variable to store this to a new variable called numNA using mutate().
We can also create a table showing how many participants have a particular number of NA values. Here we use two additional commands that you haven’t seen yet. The group_by() function tells other functions to do their analyses while breaking the data into groups based on one of the variables. Here we are going to want to summarize the number of people with each possible number of NAs, so we will group responses by the numNA variable that we are creating in the first command here.
The summarize() function creates a summary of the data, with the new variables based on the data being fed in. In this case, we just want to count up the number of subjects in each group, which we can do using the special n() function from dpylr.
# compute the number of NAs for each participant
attitudeData <-
attitudeData %>%
mutate(
numNA = rowSums(is.na(.)) # we use the . symbol to tell the is.na function to look at the entire data frame
)
# present a table with counts of the number of missing responses
attitudeData %>%
count(numNA)## # A tibble: 3 x 2
## numNA n
## <dbl> <int>
## 1 0 141
## 2 1 6
## 3 2 1We can see from the table that there are only a few participants with missing data; six people are missing one answer, and one is missing two answers. Let’s find those individuals, using the filter() command from dplyr. filter() returns the subset of rows from a data frame that match a particular test - in this case, whether numNA is > 0.
attitudeData %>%
filter(numNA > 0)## # A tibble: 7 x 33
## ID Year StatsBefore ats1 ats2 ats3 ats4 ats5
## <chr> <dbl> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 42 2 No NA 2 7 5 6
## 2 55 1 No 5 3 7 4 5
## 3 90 1 No 7 2 7 5 7
## 4 113 2 No 5 7 7 5 6
## 5 117 2 Yes 6 6 7 4 6
## 6 137 3 No 7 5 6 5 6
## 7 139 1 No 7 5 7 5 6
## # … with 25 more variables: ats6 <dbl>, ats7 <dbl>,
## # ats8 <dbl>, ats9 <dbl>, ats10 <dbl>, ats11 <dbl>,
## # ats12 <dbl>, ats13 <dbl>, ats14 <dbl>, ats15 <dbl>,
## # ats16 <dbl>, ats17 <dbl>, ats18 <dbl>, ats19 <dbl>,
## # ats20 <dbl>, ats21 <dbl>, ats22 <dbl>, ats23 <dbl>,
## # ats24 <dbl>, ats25 <dbl>, ats26 <dbl>, ats27 <dbl>,
## # ats28 <dbl>, ats29 <dbl>, numNA <dbl>There are fancy techniques for trying to guess the value of missing data (known as “imputation”) but since the number of participants with missing values is small, let’s just drop those participants from the list. We can do this using the drop_na() function from the tidyr package, another tidyverse package that provides tools for cleaning data. We will also remove the numNA variable, since we won’t need it anymore after removing the subjects with missing answeres. We do this using the select() function from the dplyr tidyverse package, which selects or removes columns from a data frame. By putting a minus sign in front of numNA, we are telling it to remove that column.
select() and filter() are similar - select() works on columns (i.e. variables) and filter() works on rows (i.e. observations).
# this is equivalent to drop_na(attitudeData)
attitudeDataNoNA <-
attitudeData %>%
drop_na() %>%
select(-numNA)
glimpse(attitudeDataNoNA)## Observations: 141
## Variables: 32
## $ ID <chr> "1", "2", "3", "4", "5", "6", "7", "8…
## $ Year <dbl> 3, 4, 2, 1, 2, 3, 4, 2, 2, 2, 4, 2, 3…
## $ StatsBefore <chr> "Yes", "No", "No", "Yes", "No", "No",…
## $ ats1 <dbl> 6, 4, 6, 3, 7, 4, 6, 5, 7, 5, 5, 4, 2…
## $ ats2 <dbl> 1, 5, 5, 2, 7, 5, 5, 4, 2, 2, 3, 3, 7…
## $ ats3 <dbl> 7, 6, 5, 7, 2, 4, 7, 7, 7, 5, 6, 5, 7…
## $ ats4 <dbl> 2, 5, 5, 2, 7, 3, 3, 4, 5, 3, 3, 2, 3…
## $ ats5 <dbl> 7, 5, 6, 7, 5, 4, 6, 6, 7, 5, 3, 5, 4…
## $ ats6 <dbl> 1, 4, 5, 2, 2, 4, 2, 3, 1, 2, 2, 3, 1…
## $ ats7 <dbl> 1, 4, 3, 2, 4, 4, 2, 2, 3, 2, 4, 2, 4…
## $ ats8 <dbl> 2, 1, 4, 3, 1, 4, 4, 4, 7, 3, 2, 4, 1…
## $ ats9 <dbl> 5, 4, 5, 5, 7, 4, 5, 5, 7, 6, 3, 5, 5…
## $ ats10 <dbl> 2, 3, 2, 2, 1, 4, 2, 2, 1, 3, 3, 1, 1…
## $ ats11 <dbl> 6, 4, 6, 2, 7, 4, 6, 5, 7, 3, 3, 4, 2…
## $ ats12 <dbl> 2, 4, 1, 2, 5, 7, 2, 1, 2, 4, 4, 2, 4…
## $ ats13 <dbl> 6, 4, 5, 5, 7, 3, 6, 6, 7, 5, 2, 5, 1…
## $ ats14 <dbl> 2, 4, 3, 3, 3, 4, 2, 1, 1, 3, 3, 2, 1…
## $ ats15 <dbl> 2, 4, 3, 3, 5, 6, 3, 4, 2, 3, 2, 4, 3…
## $ ats16 <dbl> 1, 3, 2, 5, 1, 5, 2, 1, 2, 3, 2, 2, 1…
## $ ats17 <dbl> 7, 7, 5, 7, 7, 4, 6, 6, 7, 6, 6, 7, 4…
## $ ats18 <dbl> 2, 5, 4, 5, 7, 4, 2, 4, 2, 5, 2, 4, 6…
## $ ats19 <dbl> 3, 3, 4, 3, 2, 3, 6, 5, 7, 3, 3, 5, 2…
## $ ats20 <dbl> 1, 4, 1, 2, 1, 4, 2, 2, 1, 2, 3, 2, 3…
## $ ats21 <dbl> 6, 3, 5, 5, 7, 5, 6, 5, 7, 3, 4, 6, 6…
## $ ats22 <dbl> 7, 4, 5, 6, 7, 5, 6, 5, 7, 5, 5, 5, 5…
## $ ats23 <dbl> 6, 4, 6, 6, 7, 5, 6, 7, 7, 5, 3, 5, 3…
## $ ats24 <dbl> 7, 4, 4, 6, 7, 5, 6, 5, 7, 5, 5, 5, 3…
## $ ats25 <dbl> 3, 5, 3, 3, 5, 4, 3, 4, 2, 3, 3, 2, 5…
## $ ats26 <dbl> 7, 4, 5, 6, 2, 4, 6, 5, 7, 3, 4, 4, 2…
## $ ats27 <dbl> 2, 4, 2, 2, 4, 4, 2, 1, 2, 3, 3, 2, 1…
## $ ats28 <dbl> 2, 4, 3, 5, 2, 3, 3, 1, 1, 4, 3, 2, 2…
## $ ats29 <dbl> 4, 4, 3, 6, 2, 1, 5, 3, 3, 3, 2, 3, 2…Try the following on your own: Using the attitudeData data frame, drop the NA values, create a new variable called mystery that contains a value of 1 for anyone who answered 7 to question ats4 (“Statistics seems very mysterious to me”). Create a summary that includes the number of people reporting 7 on this question, and the proportion of people who reported 7.
5.7.1.1 Tidy data
These data are in a format that meets the principles of “tidy data”, which state the following:
- Each variable must have its own column.
- Each observation must have its own row.
- Each value must have its own cell.
This is shown graphically the following figure (from Hadley Wickham, developer of the “tidyverse”):
[Following three rules makes a dataset tidy: variables are in columns, observations are in rows, and values are in cells..]
In our case, each column represents a variable: ID identifies which student responded, Year contains their year at Stanford, StatsBefore contains whether or not they have taken statistics before, and ats1 through ats29 contain their responses to each item on the ATS scale. Each observation (row) is a response from one individual student. Each value has its own cell (e.g., the values for Year and StatsBefoe are stored in separate cells in separate columns).
For an example of data that are NOT tidy, take a look at these data Belief in Hell - click on the “Table” tab to see the data.
- What are the variables
- Why aren’t these data tidy?
5.7.1.2 Recoding data
We now have tidy data; however, some of the ATS items require recoding. Specifically, some of the items need to be “reverse coded”; these items include: ats2, ats4, ats6, ats7, ats10, ats12, ats14, ats15, ats16, ats18, ats20, ats25, ats27 and ats28. The raw responses for each item are on the 1-7 scale; therefore, for the reverse coded items, we need to reverse them by subtracting the raw score from 8 (such that 7 becomes 1 and 1 becomes 7). To recode these items, we will use the tidyverse mutate() function. It’s a good idea when recoding to preserve the raw original variables and create new recoded variables with different names.
There are two ways we can use mutate() function to recode these variables. The first way is easier to understand as a new code, but less efficient and more prone to error. Specifically, we repeat the same code for every variable we want to reverse code as follows:
attitudeDataNoNA %>%
mutate(
ats2_re = 8 - ats2,
ats4_re = 8 - ats4,
ats6_re = 8 - ats6,
ats7_re = 8 - ats7,
ats10_re = 8 - ats10,
ats12_re = 8 - ats12,
ats14_re = 8 - ats14,
ats15_re = 8 - ats15,
ats16_re = 8 - ats16,
ats18_re = 8 - ats18,
ats20_re = 8 - ats20,
ats25_re = 8 - ats25,
ats27_re = 8 - ats27,
ats28_re = 8 - ats28
) ## # A tibble: 141 x 46
## ID Year StatsBefore ats1 ats2 ats3 ats4 ats5
## <chr> <dbl> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 1 3 Yes 6 1 7 2 7
## 2 2 4 No 4 5 6 5 5
## 3 3 2 No 6 5 5 5 6
## 4 4 1 Yes 3 2 7 2 7
## 5 5 2 No 7 7 2 7 5
## 6 6 3 No 4 5 4 3 4
## 7 7 4 Yes 6 5 7 3 6
## 8 8 2 Yes 5 4 7 4 6
## 9 9 2 Yes 7 2 7 5 7
## 10 10 2 Yes 5 2 5 3 5
## # … with 131 more rows, and 38 more variables: ats6 <dbl>,
## # ats7 <dbl>, ats8 <dbl>, ats9 <dbl>, ats10 <dbl>,
## # ats11 <dbl>, ats12 <dbl>, ats13 <dbl>, ats14 <dbl>,
## # ats15 <dbl>, ats16 <dbl>, ats17 <dbl>, ats18 <dbl>,
## # ats19 <dbl>, ats20 <dbl>, ats21 <dbl>, ats22 <dbl>,
## # ats23 <dbl>, ats24 <dbl>, ats25 <dbl>, ats26 <dbl>,
## # ats27 <dbl>, ats28 <dbl>, ats29 <dbl>, ats2_re <dbl>,
## # ats4_re <dbl>, ats6_re <dbl>, ats7_re <dbl>,
## # ats10_re <dbl>, ats12_re <dbl>, ats14_re <dbl>,
## # ats15_re <dbl>, ats16_re <dbl>, ats18_re <dbl>,
## # ats20_re <dbl>, ats25_re <dbl>, ats27_re <dbl>,
## # ats28_re <dbl>The second way is more efficient and takes advatange of the use of “scoped verbs” (https://dplyr.tidyverse.org/reference/scoped.html), which allow you to apply the same code to several variables at once. Because you don’t have to keep repeating the same code, you’re less likely to make an error:
ats_recode <- #create a vector of the names of the variables to recode
c(
"ats2",
"ats4",
"ats6",
"ats7",
"ats10",
"ats12",
"ats14",
"ats15",
"ats16",
"ats18",
"ats20",
"ats25",
"ats27",
"ats28"
)
attitudeDataNoNA <-
attitudeDataNoNA %>%
mutate_at(
vars(ats_recode), # the variables you want to recode
funs(re = 8 - .) # the function to apply to each variable
)Whenever we do an operation like this, it’s good to check that it actually worked correctly. It’s easy to make mistakes in coding, which is why it’s important to check your work as well as you can.
We can quickly select a couple of the raw and recoded columns from our data and make sure things appear to have gone according to plan:
attitudeDataNoNA %>%
select(
ats2,
ats2_re,
ats4,
ats4_re
)## # A tibble: 141 x 4
## ats2 ats2_re ats4 ats4_re
## <dbl> <dbl> <dbl> <dbl>
## 1 1 7 2 6
## 2 5 3 5 3
## 3 5 3 5 3
## 4 2 6 2 6
## 5 7 1 7 1
## 6 5 3 3 5
## 7 5 3 3 5
## 8 4 4 4 4
## 9 2 6 5 3
## 10 2 6 3 5
## # … with 131 more rowsLet’s also make sure that there are no responses outside of the 1-7 scale that we expect, and make sure that no one specified a year outside of the 1-4 range.
attitudeDataNoNA %>%
summarise_at(
vars(ats1:ats28_re),
funs(min, max)
)## # A tibble: 1 x 86
## ats1_min ats2_min ats3_min ats4_min ats5_min ats6_min
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 1 1 2 1 2 1
## # … with 80 more variables: ats7_min <dbl>, ats8_min <dbl>,
## # ats9_min <dbl>, ats10_min <dbl>, ats11_min <dbl>,
## # ats12_min <dbl>, ats13_min <dbl>, ats14_min <dbl>,
## # ats15_min <dbl>, ats16_min <dbl>, ats17_min <dbl>,
## # ats18_min <dbl>, ats19_min <dbl>, ats20_min <dbl>,
## # ats21_min <dbl>, ats22_min <dbl>, ats23_min <dbl>,
## # ats24_min <dbl>, ats25_min <dbl>, ats26_min <dbl>,
## # ats27_min <dbl>, ats28_min <dbl>, ats29_min <dbl>,
## # ats2_re_min <dbl>, ats4_re_min <dbl>,
## # ats6_re_min <dbl>, ats7_re_min <dbl>,
## # ats10_re_min <dbl>, ats12_re_min <dbl>,
## # ats14_re_min <dbl>, ats15_re_min <dbl>,
## # ats16_re_min <dbl>, ats18_re_min <dbl>,
## # ats20_re_min <dbl>, ats25_re_min <dbl>,
## # ats27_re_min <dbl>, ats28_re_min <dbl>, ats1_max <dbl>,
## # ats2_max <dbl>, ats3_max <dbl>, ats4_max <dbl>,
## # ats5_max <dbl>, ats6_max <dbl>, ats7_max <dbl>,
## # ats8_max <dbl>, ats9_max <dbl>, ats10_max <dbl>,
## # ats11_max <dbl>, ats12_max <dbl>, ats13_max <dbl>,
## # ats14_max <dbl>, ats15_max <dbl>, ats16_max <dbl>,
## # ats17_max <dbl>, ats18_max <dbl>, ats19_max <dbl>,
## # ats20_max <dbl>, ats21_max <dbl>, ats22_max <dbl>,
## # ats23_max <dbl>, ats24_max <dbl>, ats25_max <dbl>,
## # ats26_max <dbl>, ats27_max <dbl>, ats28_max <dbl>,
## # ats29_max <dbl>, ats2_re_max <dbl>, ats4_re_max <dbl>,
## # ats6_re_max <dbl>, ats7_re_max <dbl>,
## # ats10_re_max <dbl>, ats12_re_max <dbl>,
## # ats14_re_max <dbl>, ats15_re_max <dbl>,
## # ats16_re_max <dbl>, ats18_re_max <dbl>,
## # ats20_re_max <dbl>, ats25_re_max <dbl>,
## # ats27_re_max <dbl>, ats28_re_max <dbl>attitudeDataNoNA %>%
summarise_at(
vars(Year),
funs(min, max)
)## # A tibble: 1 x 2
## min max
## <dbl> <dbl>
## 1 1 45.7.1.3 Different data formats
Sometimes we need to reformat our data in order to analyze it or visualize it in a specific way. Two tidyverse functions, gather() and spread(), help us to do this.
For example, say we want to examine the distribution of the raw responses to each of the ATS items (i.e., a histogram). In this case, we would need our x-axis to be a single column of the responses across all the ATS items. However, currently the responses for each item are stored in 29 different columns.
This means that we need to create a new version of this dataset. It will have four columns: - ID - Year - Question (for each of the ATS items) - ResponseRaw (for the raw response to each of the ATS items)
Thus, we want change the format of the dataset from being “wide” to being “long”.
We change the format to “wide” using the gather() function.
gather() takes a number of variables and reformates them into two variables: one that contains the variable values, and another called the “key” that tells us which variable the value came from. In this case, we want it to reformat the data so that each response to an ATS question is in a separate row and the key column tells us which ATS question it corresponds to. It is much better to see this in practice than to explain in words!
attitudeData_long <-
attitudeDataNoNA %>%
select(-ats_recode) %>% #remove the raw variables that you recoded
gather(
key = question, # key refers to the new variable containing the question number
value = response, # value refers to the new response variable
-ID, -Year, -StatsBefore #the only variables we DON'T want to gather
)
attitudeData_long %>%
slice(1:20)## # A tibble: 20 x 5
## ID Year StatsBefore question response
## <chr> <dbl> <chr> <chr> <dbl>
## 1 1 3 Yes ats1 6
## 2 2 4 No ats1 4
## 3 3 2 No ats1 6
## 4 4 1 Yes ats1 3
## 5 5 2 No ats1 7
## 6 6 3 No ats1 4
## 7 7 4 Yes ats1 6
## 8 8 2 Yes ats1 5
## 9 9 2 Yes ats1 7
## 10 10 2 Yes ats1 5
## 11 11 4 No ats1 5
## 12 12 2 No ats1 4
## 13 13 3 Yes ats1 2
## 14 14 1 Yes ats1 6
## 15 15 2 No ats1 7
## 16 16 4 No ats1 7
## 17 17 2 No ats1 7
## 18 18 2 No ats1 6
## 19 19 1 No ats1 6
## 20 20 1 No ats1 3glimpse(attitudeData_long)## Observations: 4,089
## Variables: 5
## $ ID <chr> "1", "2", "3", "4", "5", "6", "7", "8…
## $ Year <dbl> 3, 4, 2, 1, 2, 3, 4, 2, 2, 2, 4, 2, 3…
## $ StatsBefore <chr> "Yes", "No", "No", "Yes", "No", "No",…
## $ question <chr> "ats1", "ats1", "ats1", "ats1", "ats1…
## $ response <dbl> 6, 4, 6, 3, 7, 4, 6, 5, 7, 5, 5, 4, 2…Say we now wanted to undo the gather() and return our dataset to wide format. For this, we would use the function spread().
attitudeData_wide <-
attitudeData_long %>%
spread(
key = question, #key refers to the variable indicating which question each response belongs to
value = response
)
attitudeData_wide %>%
slice(1:20)## # A tibble: 20 x 32
## ID Year StatsBefore ats1 ats10_re ats11 ats12_re
## <chr> <dbl> <chr> <dbl> <dbl> <dbl> <dbl>
## 1 1 3 Yes 6 6 6 6
## 2 10 2 Yes 5 5 3 4
## 3 100 4 Yes 5 6 4 2
## 4 101 2 No 4 7 2 4
## 5 102 3 Yes 5 6 5 6
## 6 103 2 No 6 7 5 7
## 7 104 2 Yes 6 5 5 3
## 8 105 3 No 6 6 5 6
## 9 106 1 No 4 4 4 4
## 10 107 2 No 1 2 1 1
## 11 108 2 No 7 7 7 7
## 12 109 2 No 4 4 4 6
## 13 11 4 No 5 5 3 4
## 14 110 3 No 5 7 4 4
## 15 111 2 No 6 6 6 3
## 16 112 3 No 6 7 5 7
## 17 114 2 No 5 4 4 3
## 18 115 3 No 5 7 5 1
## 19 116 3 No 5 6 5 5
## 20 118 2 No 6 6 6 1
## # … with 25 more variables: ats13 <dbl>, ats14_re <dbl>,
## # ats15_re <dbl>, ats16_re <dbl>, ats17 <dbl>,
## # ats18_re <dbl>, ats19 <dbl>, ats2_re <dbl>,
## # ats20_re <dbl>, ats21 <dbl>, ats22 <dbl>, ats23 <dbl>,
## # ats24 <dbl>, ats25_re <dbl>, ats26 <dbl>,
## # ats27_re <dbl>, ats28_re <dbl>, ats29 <dbl>,
## # ats3 <dbl>, ats4_re <dbl>, ats5 <dbl>, ats6_re <dbl>,
## # ats7_re <dbl>, ats8 <dbl>, ats9 <dbl>Now that we have created a “long” version of our data, they are in the right format to create the plot. We will use the tidyverse function ggplot() to create our histogram with geom_histogram.
attitudeData_long %>%
ggplot(aes(x = response)) +
geom_histogram(binwidth = 0.5) +
scale_x_continuous(breaks = seq.int(1, 7, 1))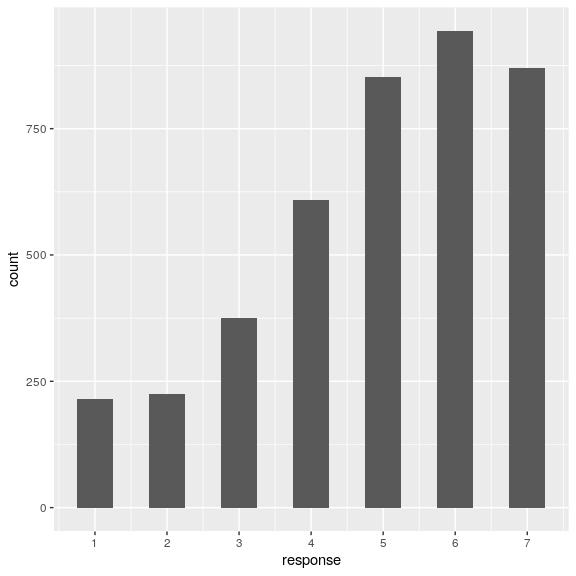
It looks like responses were fairly positively overall.
We can also aggregate each participant’s responses to each question during each year of their study at Stanford to examine the distribution of mean ATS responses across people by year.
We will use the group_by() and summarize() functions to aggregate the responses.
attitudeData_agg <-
attitudeData_long %>%
group_by(ID, Year) %>%
summarize(
mean_response = mean(response)
)
attitudeData_agg## # A tibble: 141 x 3
## # Groups: ID [141]
## ID Year mean_response
## <chr> <dbl> <dbl>
## 1 1 3 6
## 2 10 2 4.66
## 3 100 4 5.03
## 4 101 2 5.10
## 5 102 3 4.66
## 6 103 2 5.55
## 7 104 2 4.31
## 8 105 3 5.10
## 9 106 1 4.21
## 10 107 2 2.45
## # … with 131 more rowsFirst let’s use the geom_density argument in ggplot() to look at mean responses across people, ignoring year of response. The density argrument is like a histogram but smooths things over a bit.
attitudeData_agg %>%
ggplot(aes(mean_response)) +
geom_density()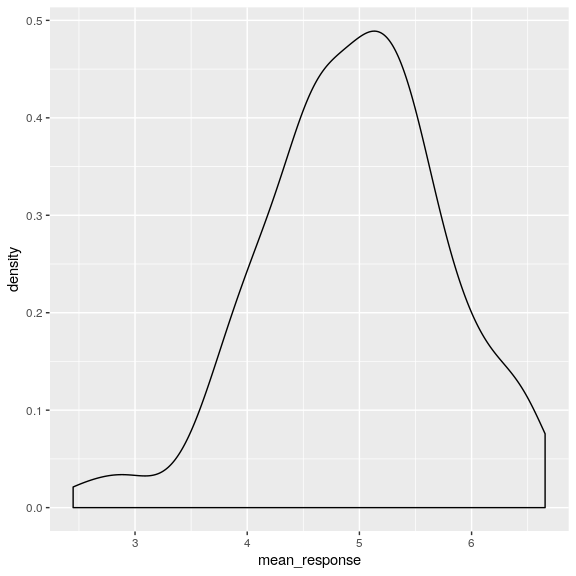
Now we can also look at the distribution for each year.
attitudeData_agg %>%
ggplot(aes(mean_response, color = factor(Year))) +
geom_density()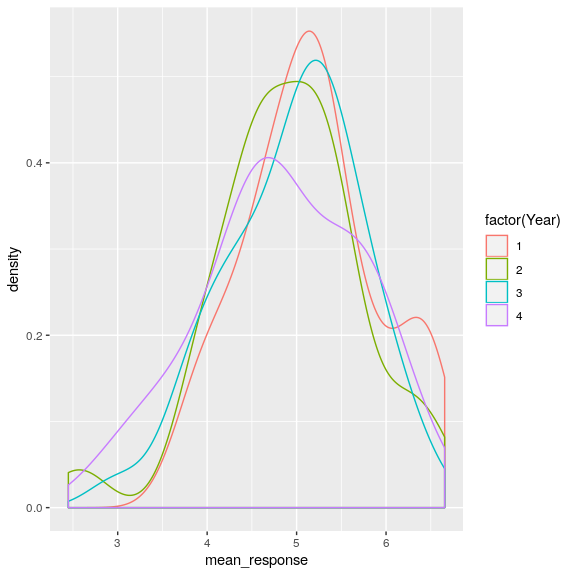
Or look at trends in responses across years.
attitudeData_agg %>%
group_by(Year) %>%
summarise(
mean_response = mean(mean_response)
) %>%
ggplot(aes(Year, mean_response)) +
geom_line()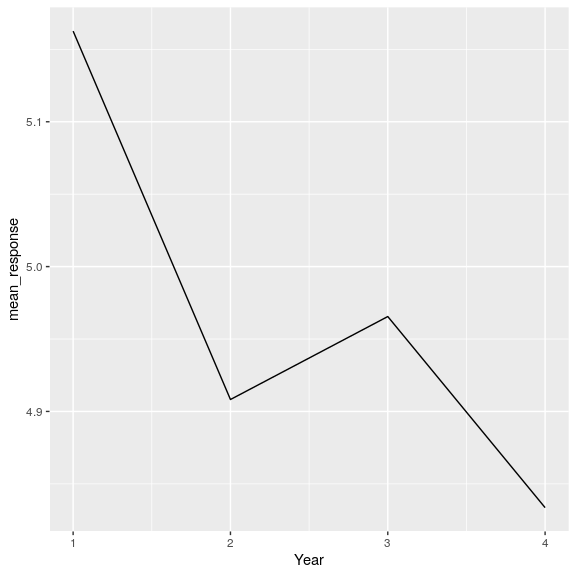
This looks like a precipitous drop - but how might that be misleading?


