8.5: One sample t-test
- Page ID
- 45186
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Introduction
We’re now talking about the traditional, classical two-group comparison involving continuous data types. Thus begins your introduction to parametric statistics. One-sample tests involve questions like “how many (what proportion of) people would we expect are shorter or taller than two standard deviations from the mean?” This type of question assumes a population and we use properties of the normal distribution and, hence, these are called parametric tests because the assumption is that the data has been sampled from a particular probability distribution.
However, when we start asking questions about a sample statistic (e.g., the sample mean), we cannot use the normal distribution directly, i.e., we cannot use \(Z\) and the normal table as we did before (Chapter 6.7). This is because we do not know the population standard deviation and therefore must use an estimate of the variation \((s)\) to calculate the standard error of the mean.
With the introduction of the \(t\)-statistic, we’re now into full inferential statistics-mode. What we do have are estimates of these parameters. The \(t\)-test — aka Student’s t-test — was developed for the purpose of testing sample means when the true population parameters are not known.
It’s called Student’s \(t\)-test after the pseudonym used by William Gosset.
This is the equation of the one sample t-test. Note the resemblance in form with the Z-score! \[t = \frac{\bar{X} - \mu}{s_{\bar{X}}} \nonumber\]
where \(s_{\bar{X}}\) is the sample standard error of the sample mean (SEM).
For example, weight change of mice given a hormone (leptin) or placebo. \(\bar{X} = 5 \text{ g}\), but under the null hypothesis, the mean change is “really” zero \(\mu = 0\). How unlikely is our value of 5 grams?
Notice how I snuck in “placebo” and mice? Do you think the concept of placebo is appropriate for research with mice, or should we simply refer to it as a control treatment? See Ch. 5.4 – Clinical trials for review.
Speaking of null hypotheses, can you say (or write) the null and alternative hypotheses in this example? How about in symbolic form?
We want to know if our sample mean could have been obtained by chance alone from a population where the true change in weight was zero.
\(s = 3\), \(n = 20\), and \(s_{X} = \dfrac{s}{\sqrt{n}} = \dfrac{3}{\sqrt{20}} = 0.6708\)
We take these values and plug them into our equation of the \(t\)-test: \[t = \frac{5 - 0}{0.67} = 7.45 \nonumber\]
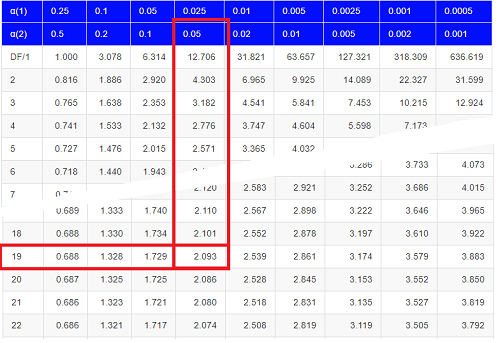
Then recall that Degrees of Freedom are \(DF = n - 1\), so we have \(DF = 20 - 1 = 19\) for the one sample \(t\)-test. And the Critical Value is found in the appropriate table of critical values for the \(t\) distribution (Fig. \(\PageIndex{1}\)).
Or, and better, use R
qt(c(0.025), df=19, lower.tail=FALSE)
where qt() is function call to find t-score of the pth percentile (cf 3.3 – Measures of dispersion) of the Student \(t\) distribution. For a two-tailed test, we recall that 0.025 is lower tail and 0.025 is upper tail.
In this example we would be willing to reject the Null Hypothesis if there was a positive OR a negative change in weight.
This was an example of a “two-tailed test,” which is “2-tail” or \(\alpha_{(2)}\) in the table of critical values of the \(t\) distribution.
The critical value for \(\alpha_{(2)} = 0.05\) and \(df = 19\) is 2.093. Do we accept or reject the Null Hypothesis?
A typical inference workflow
Note the general form of how the statistical test is processed, a form which actually applies to any statistical inference test.
- Identify the type of data
- State the null hypothesis (2-tailed? 1-tailed?)
- Select the test statistic (\(t\)-test) and determine its properties
- Calculate the test statistic (the value of the result of the \(t\)-test)
- Find degrees of freedom
- For the DF, get the critical value
- Compare critical value to test statistic
- Do we accept or reject the null hypothesis?
And then we ask, given the results of the test of inference, What is the biological interpretation? Statistical significance is not necessarily evidence of biological importance. In addition to statistical significance, the magnitude of the difference — the effect size — is important as part of interpreting results from an experiment. Statistical significance is at least in part because of sample size — the large the sample size, the smaller the standard error of the mean, therefore even small differences may be statistically significant, yet biologically unimportant. Effect size is discussed in Ch. 9.1 – Chi-square test: Goodness of fit, Ch. 11.4 – Two-sample effect size and Ch. 12.5 – Effect size for ANOVA.
R Code
Let’s try a one-sample \(t\)-test. Consider the following data set: body mass of four geckos and four Anoles lizards (Dohm unpublished data).
For starters, let’s say that you have reason to believe that the true mean for all small lizards is 5 grams (g).
Geckos: 3.186, 2.427, 4.031, 1.995 Anoles: 5.515, 5.659, 6.739, 3.184
Get the data into R (Rcmdr)
By now you should be able to load this data in one of several ways. If you haven’t already entered the data, check out Part 07. Working with your own data in Mike’s Workbook for Biostatistics.
Once we have our data.frame, proceed to carry out the statistical test.
To get the one-sample t-test in Rcmdr, click on Statistics → Means → Single-sample t-test… Because there is only one numerical variable, Body.mass, that is the only one that shows up in the Variable (pick one) window (Fig. \(\PageIndex{2}\))
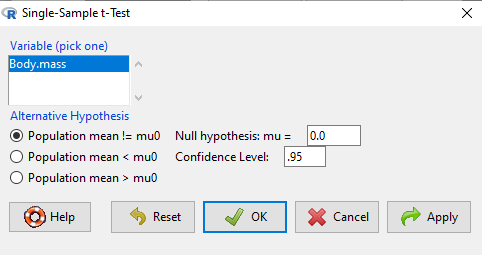
Type in the value 5.0 in the Null hypothesis: mu = box.
Question 3: Quick! Can you write, in plain old English, the statistical null hypothesis???
Click OK
The results go to the Output Window.
t.test(lizards$Body.mass, alternative = 'two.sided', mu = 5.0, conf.level = .95)
One Sample t-test data: lizards$Body.mass t = -1.5079, df = 7, p-value = 0.1753 alternative hypothesis: true mean is not equal to 5 95 percent confidence interval: 2.668108 5.515892 sample estimates: mean of x 4.092
Let’s identify the parts of the R output from the one sample t-test. R reports the name of the test and identifies:
- The dataset variable used (
lizards$Body.mass). The data set was called “lizards” and the variable was “Body.mass”. R uses the dollar sign ($) to denote the data set and variable within the data set. - The value of the statistic was \(t = -1.5079\). It is negative because the sample mean was less than the population mean — you should be able to verify this!
- The degrees of freedom: \(df = 7\)
- The p-value = 0.1753
- Confidence level = 95%
- The sample mean = 4.092
Take a step back and review
Let's make sure we “get” the logic of the hypothesis testing we have just completed. Consider the one-sample \(t\)-test.
Step 1. Define \(H_{O}\) and \(H_{A}\). The null hypothesis might be that a sample mean equals 5. \(H_{O}: \bar{X} = 5\)
The alternate is that the sample mean is not equal to 5. \(H_{A}: \bar{X} \neq 5\)
Where did the value 5 come from? It could be a value from the literature (does the new sample differ from values obtained in another lab?). The point is that the value is known in advance, before the experiment is conducted, and that makes it a one-sample \(t\)-test.
One-tailed hypothesis or two?
We introduced you to the idea of "tails of a test" (Ch. 8.4). As you should recall, a null/alternate hypothesis for a two-tailed test may be written as
\(H_{O}: \bar{X} = \mu\)
\(H_{A}: \bar{X} \neq \mu\)
Alternatively, we can write a one-tailed test null/alternate hypothesis as
\(H_{O}: \bar{X} < \mu\)
\(H_{A}: \bar{X} \geq \mu\)
Question 4: Are all possible outcomes of the one-tailed test covered by these hypotheses?
Question 5: What is the SEM for this problem?
Question 6: What is the difference between a one-sample \(t\)-test and a one-sided \(t\)-test?
Question 7: What are some other possible hypotheses that can be tested from this simple example of two lizard species?
Step 2. Decide how certain you wish to be (with what probability) that the sample mean is different from 5. As stated previously, in biology we say that we are willing to be incorrect.
Step 3. Carry out the calculation of the test statistic. In other words, get the value of \(t\) from the equation above by hand, or, if using R (yes!) simply identify the test statistic value from the R output after conducting the one-sample t-test.
Step 4. Evaluate the result of the test. If the value of the test statistic is greater than the critical value for the test, then you conclude that the chance (the P-value) that the result could be from that population is not likely and you therefore reject the null hypothesis.
Question 8: What is the critical value for a one-sample t-test with \(df = 7\)? Hint: you need the table, or better, R: Rcmdr: Distributions → Continuous distributions → t distributions → t quantiles. You also need to know three additional things to answer this question.
- You need to know \(\alpha\), which we have said is generally set at 5.
- You also need to know the degrees of freedom (DF) for the test. For a one-sample test, \(DF = n - 1\), where \(n\) is the sample size.
- You also must know whether your test is one- or two-tailed.
You then use the \(t\)-distribution (the tables of the \(t\)-distribution at the end of your book) to obtain the critical value. Note that if you use R the actual p-value is returned.
Why learn the equations when I can just do this in R?
Rcmdr does this for you for you as soon as you click OK. Rcmdr returns the value of the test statistic and the p-value. R does not show you the critical value, but instead returns the probability that your test statistic is as large as it is AND the null hypothesis is true.
The simple answer is that in order to understand the R output properly you need to know where each item of the output for a particular test comes from and how to interpret it. Thus, the best way is to have the equations available and to understand the algorithmic approach to stastical inference.
Also, this is as good of a time as any to show you how to skip the Rcmdr GUI and go straight to R.
First, create your variables. At the R prompt enter the first variable:
liz <- c("G", "G", "G", "G", "A", "A", "A", "A")
and then create the second variable:
bm <- c(3.186, 2.427, 4.031, 1.995, 5.515, 5.659, 6.739, 3.184)
Next, create a data frame. Think of a data frame as another word for worksheet.
lizz <- data.frame(liz, bm)
Verify that entries are correct. At the R prompt type “lizz” without the quotes and you should see
liz bm 1 G 3.186 2 G 2.427 3 G 4.031 4 G 1.995 5 A 5.515 6 A 5.659 7 A 6.739 8 A 3.184
Carry out the t-test by typing the following at the R prompt:
t.test(lizz, bm, alternative='two-sided', mu=5, conf.level=.95)
And, like the Rcmdr output, we have for the one-sample \(t\)-test the following R output:
One Sample t-test data: lizards$Body.mass t = -1.5079, df = 7, p-value = 0.1753 alternative hypothesis: true mean is not equal to 5 95 percent confidence interval: 2.668108 5.515892 sample estimates: mean of x 4.092
End of R output
which, as you probably guessed, is the same as what we got from RCmdr.
Question 9: From the R output of the one sample t-test, what was the value of the test statistic?
- -1.5079
- 7
- 0.1753
- 2.668108
- 5.515892
- 4.092
Note. On an exam you will be given portions of statistical tables and output from R. Thus you should be able to evaluate statistical inference questions by completing the missing information. For example, if I give you a test statistic value, whether the test is one- or two-tailed, degrees of freedom, and the Type I error rate alpha, you should know that you would need to find the critical value from the appropriate statistical table. On the other hand, if I give you R output, you should know that the p-value and whether it is less than the Type I error rate of alpha would be all that you need to answer the question.
Think of this as a basic skill.
In statistics and for some statistical tests, Rcmdr and other software may not provide the information needed to decide that your test statistic is large, and a table in a statistics book is the best way to evaluate the test.
For now, double check Rcmdr by looking up the critical value from the \(t\)-table.
Check critical value against our test statistic
\(Df = 8 - 1 = 7\)
The test is two-tailed, therefore \(\alpha(_{2})\).
\(\alpha = 0.05\) (note that two-tailed critical value is 2.365. \(t\) was equal to 1.51 (since \(t\)-distribution is symmetrical, we can ignore the negative sign), which is smaller than 2.365 and so we would agree with Rcmdr — we cannot reject the null hypothesis.
Question 10: From the R output of the one sample t-test, what was the P-value?
- -1.5079
- 7
- 0.1753
- 2.668108
- 5.515892
Question 11: We would reject the null hypothesis
- False
- True
Questions
Eleven questions were provided for you within the text in this chapter. Here’s one more.
Question 12. Here’s a small data set for you to try your hand at the one-sample \(t\)-test and Rcmdr. The dataset contains cell counts, five counts of the numbers of beads in a liquid with an automated cell counter (Scepter, Millipore USA). The true value is 200,000 beads per milliliter fluid; the manufacturer claims that the Scepter is accurate within 15%. Does the data conform to the expectations of the manufacturer? Write a hypothesis then test your hypothesis with the one-sample \(t\)-test. Here’s the data.
| Scepter |
| 258900 |
| 230300 |
| 107700 |
| 152000 |
| 136400 |


