3.2: Assumptions and Diagnostics
- Page ID
- 33434
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Before we draw any conclusions about the significance of the model, we need to make sure we have a "valid" model. Like any other statistical procedure, the ANOVA has assumptions that must be met. Failure to meet these assumptions means any conclusions drawn from the model are not to be trusted.
Assumptions
So what are these assumptions being made to employ the ANOVA model? The errors are assumed to be independent and identically distributed (iid) with a normal distribution having a mean of 0 and unknown equal variance.
As the model residuals serve as estimates of the unknown error, diagnostic tests to check for validity of model assumptions are based on residual plots, and thus, the implementation of diagnostic tests is also called Residual Analysis.
Diagnostic Tests
Most useful is the residual vs. predicted value plot, which identifies the violations of zero mean and equal variance. Residuals are also plotted against the treatment levels to examine if the residual behavior differs among treatments.
The normality assumption is checked by using a normal probability plot.
Residual plots can help identify potential outliers, and the pattern of residuals vs. fitted values or treatments may suggest a transformation of the response variable.
Lesson 4: SLR Model Assumptions of STAT 501 online notes discuss various diagnostic procedures in more detail.
There are various statistical tests to check the validity of these assumptions, but some may not be that useful. For example, Bartlett’s test for homogeneity is too sensitive and indicates that problems exist when they really don’t. It turns out that the ANOVA is very robust and is not badly affected by minor violations of these assumptions. In practice, a good deal of common sense and the visual inspection of the residual plots are sufficient to determine if serious problems exist.
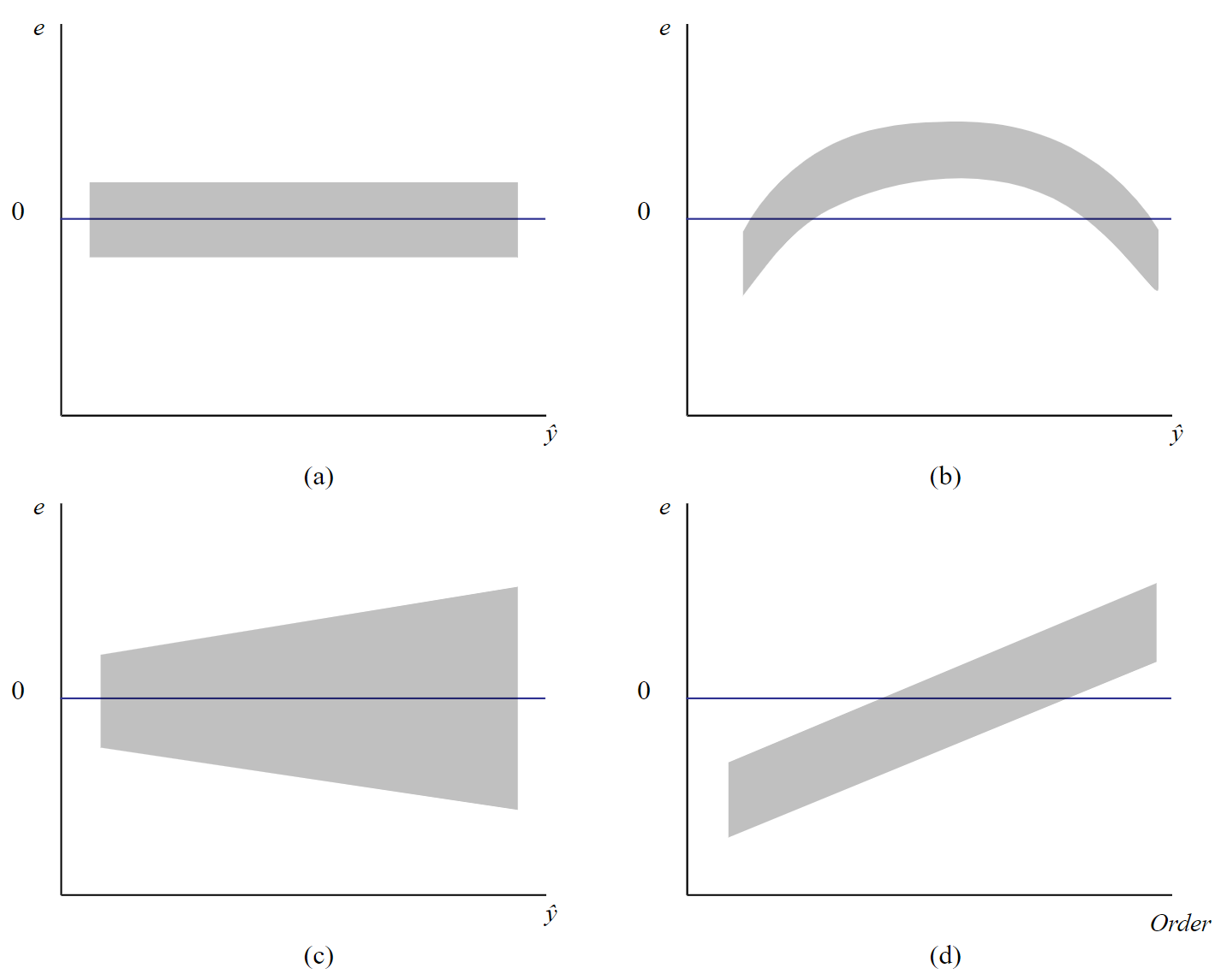
We will employ statistical software such as SAS to conduct the residual analysis. Here are common patterns that you may encounter in the residual analysis (i.e. plotting residuals, \(e\), against the predicted values, \(\hat{y}\)).
Figure \(\PageIndex{1a}\) shows the prototype plot when the ANOVA model is appropriate for data. The residuals are scattered randomly around mean zero and variability is constant (i.e. within the horizontal bands) for all groups.
.png?revision=1&size=bestfit&width=468&height=376)
Figure \(\PageIndex{1b}\) suggests that although the variance is constant, there are some trends in the response that is not explained by a linear model. Using Figure \(\PageIndex{1c}\), we can depict that the linear model is appropriate as the central trend in data is a line. However, the megaphone patterns in Figure \(\PageIndex{1c}\) suggest that variance is not constant.
A common problem encountered in ANOVA is when the variance of treatment levels is not equal (heterogeneity of variance). If the variance is increasing in proportion to the mean (panel (c) in Figure \(\PageIndex{1}\)), a logarithmic transformation of Y can "stabilize" the variances. If the residuals vs. predicted values instead show a curvilinear trend (panel (b) in Figure \(\PageIndex{1}\)), then a quadratic or other transformation may help. Since finding the correct transformation can be challenging, the Box-Cox method is often used to identify the appropriate transformation, given in terms of \(\lambda\) as shown below.
\[y_{i}^{(\lambda)} = \begin{cases} \frac{y_{i}^{\lambda} - 1}{\lambda}, \text{ if } \lambda \neq 0, \\ \ln y_{i}, \text{ if } \lambda = 0 \end{cases}\]
Some \(\lambda\) values result some common transformations.
transformations.
| \(\lambda\) | \(Y^{\lambda}\) | Transformation |
|---|---|---|
| 2 | \(Y^{2}\) | Square |
| 1 | \(Y^{1}\) | Original (No transform) |
| 1/2 | \(\sqrt{Y}\) | Square Root |
| 0 | \(\log (Y)\) | Logarithm |
| -1/2 | \(\frac{1}{\sqrt{Y}}\) | Reciprocal Square Root |
| -1 | \(\frac{1}{Y}\) | Reciprocal |
Using Technology
To run the Box-Cox procedure in Minitab, set up the data (Simulated Data), as a stacked format (a column with treatment (or trt combination) levels, and the second column with the response variable.
| Treatment | Response Variable |
|---|---|
| A | 12 |
| A | 23 |
| A | 34 |
| B | 45 |
| B | 56 |
| B | 67 |
| C | 14 |
| C | 25 |
| C | 36 |
- Steps in Minitab
-
- On the Minitab toolbar, choose Stat > Control Charts > Box-Cox Transformation
.png?revision=1&size=bestfit&width=565&height=398)
Figure \(\PageIndex{2}\): Selecting Box-Cox Transformation stat option. - Place "Response Variable" and "Treatment" in the boxes as shown below.
.png?revision=1&size=bestfit&width=720&height=324)
Figure \(\PageIndex{3}\): Inputting "Response Variable" and "Treatment" in pop-up window. - Click OK to finish. You will get an output like this:
.png?revision=1&size=bestfit&width=527&height=369)
Figure \(\PageIndex{4}\): Minitab Box-Cox plot output.
In the upper right-hand box, the rounded value for \(\lambda\) is given from which the appropriate transformation of the response variable can be found using the chart above. Note, with a \(\lambda\) of 1, no transformation is recommended.
- On the Minitab toolbar, choose Stat > Control Charts > Box-Cox Transformation
The Box-Cox procedure in SAS is more complicated in a general setting. It is done through the Transreg procedure, by obtaining the ANOVA solution with regression which first requires coding the treatment levels with effect coding discussed in Chapter 4.
However, for one-way ANOVA (ANOVA with only one factor) we can use the SAS Transreg procedure without much hassle.
- Steps in SAS
-
Suppose we have SAS data as follows.
Obs Treatment ResponseVariable 1 A 12 2 A 23 3 A 34 4 B 45 5 B 56 6 B 67 7 C 14 8 C 25 9 C 36 We can use the following SAS commands to run the Box-Cox analysis.
proc transreg data=boxcoxSimData; model boxcox(ResponseVariable)=class(Treatment); run;
This would generate an output as follows, which suggests a transformation using \(\lambda=1\) (i.e. no transformation).
.png?revision=1&size=bestfit&width=664&height=508)
Figure \(\PageIndex{5}\): SAS Box-cox plot output.
- Steps in R
-
Load the simulated data and perform the Box-Cox transformation. Note that simulated data are in the stacked format (a column with treatment levels and a column with the response variable)
setwd("~/path-to-folder/) simulated_data<-read.table("simulated_data.txt",header=T) attach(simulated_data) library(AID)#Load package AID so that we can use the Box-Cox Procedure boxcoxfr(Response_Variable,Treatment)#Box-Cox command for One-Way ANOVAOutput
Box-Cox power transformation data: Response_Variable and Treatment lambda.hat: 0.93 Shapiro-Wilk normality test for transformed data (alpha = 0.05) Level statistic p.value Normality 1 A 0.9998983 0.9807382 YES 2 B 0.9999840 0.9923681 YES 3 C 0.9999151 0.9824033 YES Bartlett's homogeneity test for transformed data (alpha = 0.05) Level statistic p.value Homogeneity 1 All 0.008271728 0.9958727 YES From the output, we can see that the lambda value for the transformation is 0.93 (the same value as Minitab suggested). Since this value is very close to 1 we can use \(\lambda=1\) (no transformation).
In addition, from the output, we can see that normality exists in all 3 levels (Shapiro-Wilk test) and we have the same variance (Bartlett's test).
Alternative:
We can use the command
boxcoxfrom package MASSlibrary(MASS) Box_Cox_Plot<-boxcox(aov(Response_Variable~Treatment),lambda=seq(-3,3,0.01))
.png?revision=1&size=bestfit&width=675&height=385)
Figure \(\PageIndex{6}\): R-generated plot of log-likelihood vs \(\lambda\). From the plot, we can see the 95% CL. Since \(\lambda=1\) is within the interval there is no need for transformation.
#Exact lambda lambda<-Box_Cox_Plot$x[which.max(Box_Cox_Plot$y)] #0.93 detach(simulated_data)


